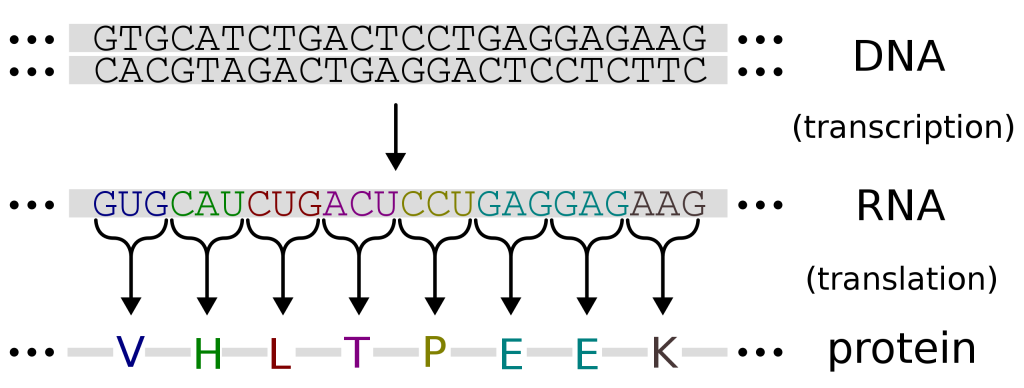
4.3 The flow of biological information
Information flows from genes to proteins but not in reverse.
The gene expression factory
You can think of a cell as a factory. In the middle of the factory is the head office (nucleus) with the instructions to make everything in the factory, including the machinery (proteins), workers (proteins), and even some other rooms (organelles). When something new needs to be made, the big manual of instructions (DNA) in the head office is sorted through and the specific instructions (one or more genes) are found. The instructions are not allowed to leave the head office, so must be photocopied (transcribed) into a single copy (messenger RNA or mRNA). This single copy leaves the head office, heads out to the factory floor (cytoplasm) and is read by a machine (ribosome). The ribosome translates the photocopy into a different language entirely – a chain (of amino acids) – and when this translation is finished the chain can contort into a new shape and becomes a functioning piece of machinery or worker that might be used in the factory or in a neighbouring factory (exported) or back in the head office (nucleus).
This analogy explains gene expression, when we go from the original gene to a functional ‘product’. Watch this video for a detailed visualisation of the process from DNA to protein.
From DNA to protein – 3D
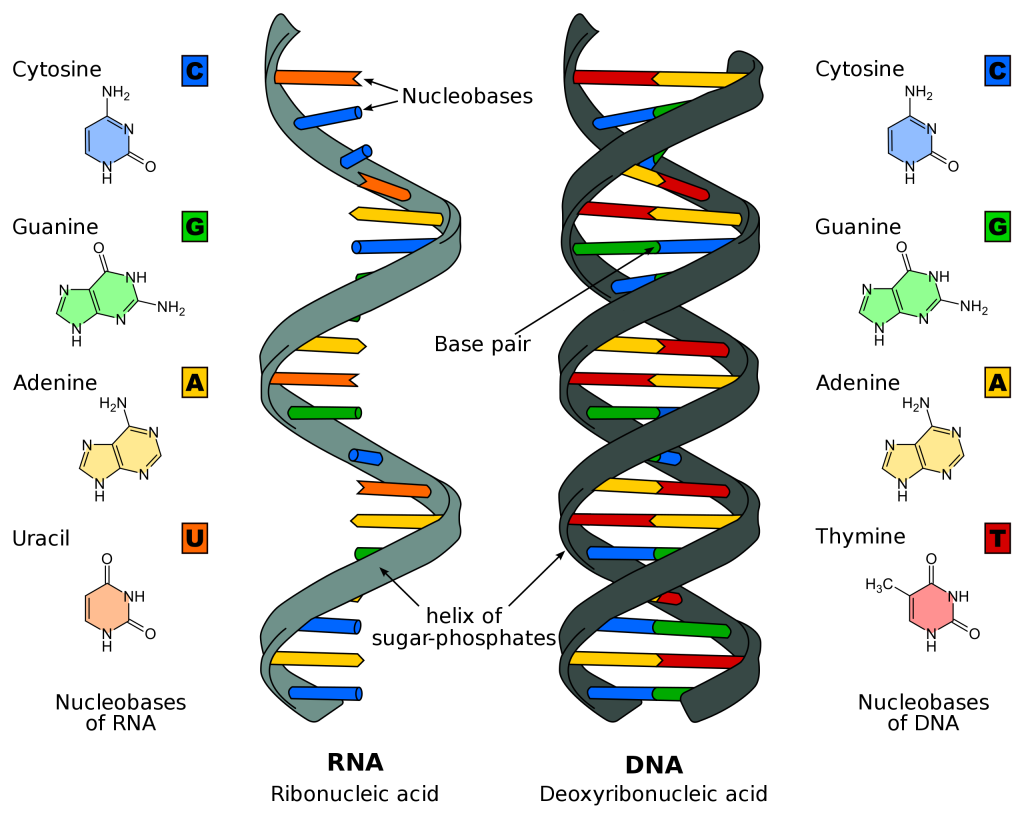
DNA vs RNA
There are two main differences between deoxyribonucleic acid (DNA) and ribonucleic acid (RNA):
- DNA is normally double stranded and RNA is normally single stranded (in humans anyway; other organisms are different).
- DNA is made up of adenine (A), thymine (T), cytosine (C) and guanine (G) bases. Each base is attached to a phosphate and a sugar molecule, which together make a nucleotide. RNA is made up of A, C and G nucleotides and instead of thymine, it has uracil (U).
In DNA, A always pairs with T, and C always pairs with G (the straight letters stick together and the curvy letters stick together) to create a base pair. In RNA, A always pairs with U. These pairings are how complementary strands ‘stick together’ and how by looking at only one strand you can replicate the DNA (if you want to pass it onto a daughter cell) or transcribe it into RNA.
Transcription
There is different machinery involved in the process from DNA to protein. During transcription, RNA polymerase finds the gene that needs to be transcribed and unwinds (‘unzips’) the double DNA strand at this location (Figure 4.4). The RNA polymerase reads the template strand of the DNA and brings in the necessary nucleotides to build a complementary mRNA strand (with uracil instead of thymine) (Figure 4.5A). When it has finished transcribing the gene (following cues from within the DNA code), the RNA is released and the RNA polymerase ‘zips’ the double-stranded DNA back together. The mRNA requires some processing before it can leave the nucleus: introns (segments of DNA or RNA that do not code for proteins) are removed through splicing, the poly-A tail (a long sequence of ‘AAAAAA’) is added to the 3’ end (three prime end) to increase the stability of the molecule and a methylated ‘cap’ is added to the 5’ end (five prime end) to help initiate protein synthesis and protect the mRNA from being degraded. In the opposite of what you might think from the names, introns are removed and exons (segments of DNA or RNA containing coding information) stay in, because intron is derived from ‘intragenic’ (inside a gene) and exons are ‘expressed’.
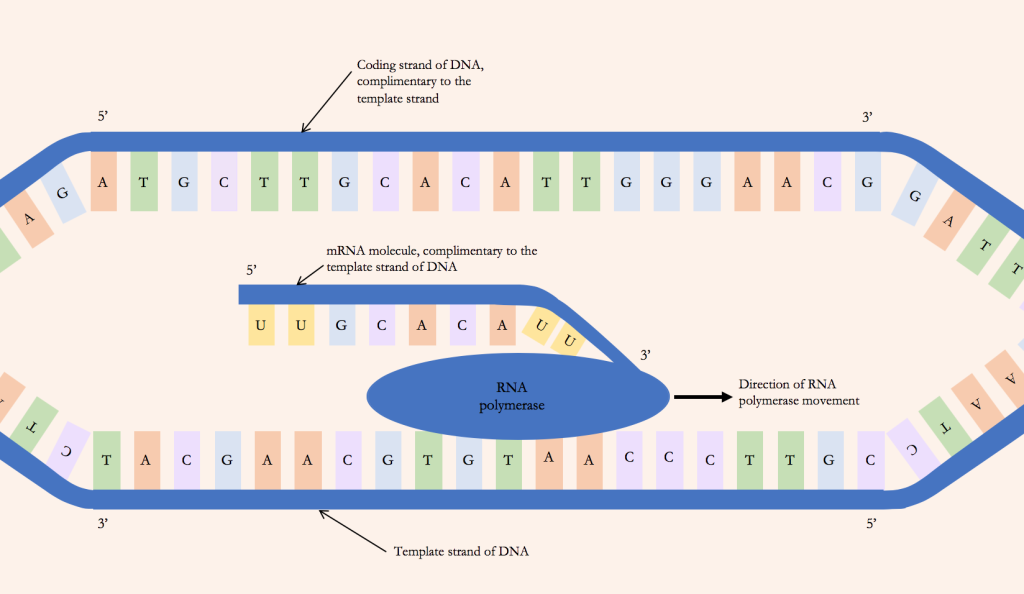
Perhaps confusingly, the ‘template’ strand of the DNA is called the non-coding strand, and the non-template strand is called the coding strand. This is because the DNA non-template strand will have the same sequence as the mRNA strand transcribed from this gene, except T’s will be U’s. In Figure 4.4 you can see that the mRNA molecule has the same sequence (apart from U/T) as the top coding strand, and these are both complementary to the template strand.
‘Transcription’ outside of biology means to convert audio or speech into written text in the same language. Therefore, as DNA and RNA use the same ‘language’ of base pairs, this process is called transcription.
Translation
Now that the ‘photocopy’ of the gene has been made, the mRNA can leave the nucleus through a little pore and find a ribosome. The ribosome reads the RNA in codons (three nucleotides in a row) and recruits transfer RNA (tRNA) with a complementary anti-codon to the RNA sequence. The tRNAs have a specific anti-codon at their ‘feet’, and carry a specific amino acid on their ‘head’. Within the ribosome, when the tRNA anti-codon matches up with the codon in the mRNA the amino acid is released from the head of the tRNA and joins the growing amino acid chain. This process continues until all the mRNA codons have been matched with tRNA anti-codons and a ‘stop’ codon is reached; then the amino acid chain is complete. This amino acid chain may then undergo further processing, fold into a certain shape or join with other amino acid subunits to make a functional protein. Note that some genes do not undergo translation, they are destined to remain as RNA such as ribosomal or transfer RNAs (see Figure 4.5 B, C).
As the RNA and amino acids are in different ‘languages’, this process is called translation.
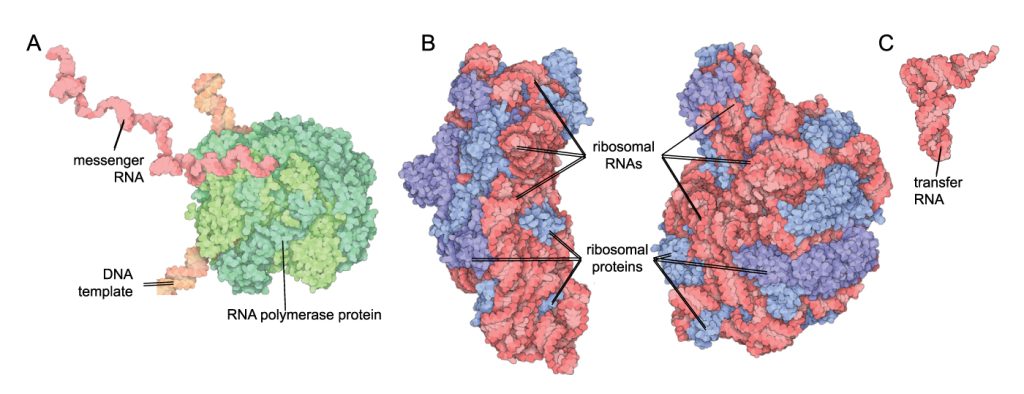
Translation is decoding RNA into protein.
The central dogma of molecular biology
The central dogma of molecular biology is an explanation of the flow of genetic information in a biological system. It was first stated by Francis Crick, who together with James Watson and Maurice Wilkins was awarded a Nobel prize in 1962 for explaining the helical structure of DNA. The dogma is often stated as:
DNA is transcribed to RNA which is translated to protein.
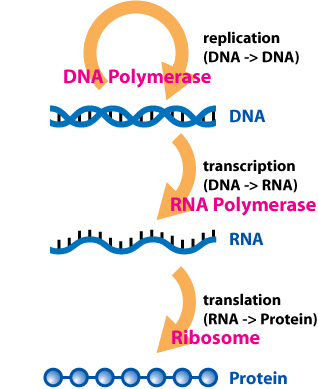
Depicting the one-way flow of biological information from DNA to RNA to protein and identifying the key enzymes and machinery involved at each step. Image: ‘Central Dogma of Molecular Biochemistry with Enzymes’ by Daniel Horspool from Wikimedia Commons used under CC BY-SA 3.0.
Decoding: understanding the triplet code
As we’ve explained, the four little letters of the DNA alphabet encode all life. Ribosomes read the mRNA in groups of three nucleotides called codons, and the specific order of the nucleotides in the codon determine which amino acid joins the chain. The amino acid codon table (Figure 4.7) shows how to determine which amino acid is encoded for by the codon in the mRNA. The next video illustrates how to use this table.
How to read a codon chart
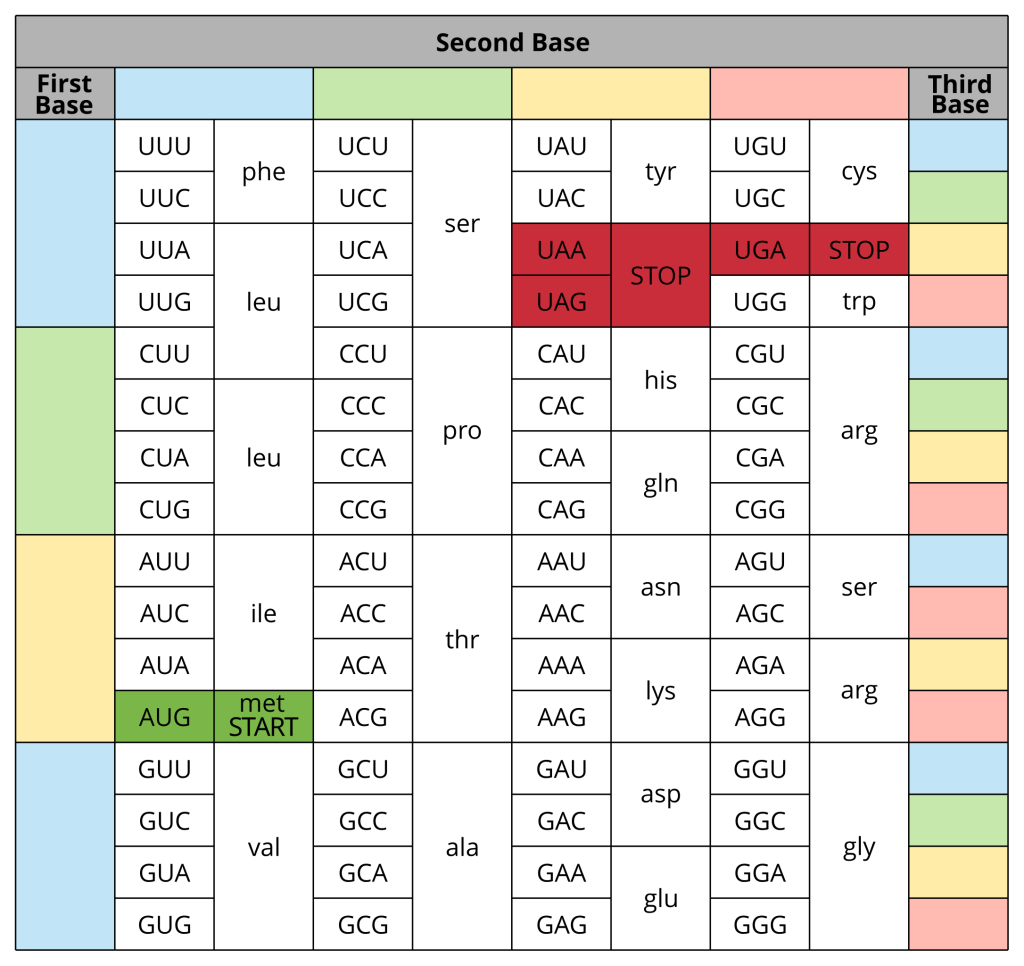
There are four important codons highlighted in the table to keep in mind:
- AUG is known as the start codon. Ribosomes know to look for it as it signals where to start translating the mRNA and also establishes the reading frame. AUG encodes for methionine, which is almost always the first amino acid in the chain but can also be inserted in other locations along the chain.
- UAA, UAG and UGA are known as stop codons. They do not encode for an amino acid and instead at this position the amino acid strand will be released from the ribosome and translation is complete.
Codon redundancy
Studying the amino acid codon table (Figure 4.7) we can see that there is some redundancy – for example, UCU, UCC, UCA and UCG all encode for the amino acid serine, while CGU, CGC, CGA, CGG, AGA and AGG encode for arginine. This makes sense. There are only 20 amino acids, but 64 possible combinations of the four nucleotides (4 × 4 × 4). It’s remarkable that all life uses this same coding system to build proteins from the genes we inherit.
This redundancy is also useful if there is a mutation at the third base of a codon (e.g. a C mutated to an A in UCC) – the amino acid encoded would not be different; we would still get serine added to the amino acid chain. You might think a single amino acid change isn’t too much of an issue, but it could have detrimental effects. A single change could alter the shape of the final protein, alter the active site of an enzyme or change the hydrophobic/hydrophilic nature of the protein so that it no longer sits in the plasma membrane. This might not sound problematic, but if the enzyme has an important job or the plasma membrane protein pumps a molecule that your cell can’t live without then a small amino acid change could be fatal. Imagine if you had another accelerator pedal in your car instead of a brake. It looks like a trivial change (you have the same number of pedals), but it’s likely to be seriously detrimental. It is important to keep in mind that a lot of mutations in DNA code are not ‘solved’ by this codon redundancy though.
Limitations to information flow
As we’ve discussed, DNA is transcribed into RNA and RNA is translated into protein (figures 4.8 and 4.9). If you were given a string of DNA letters, you would be able to transcribe this yourself into RNA (knowing what bases/nucleotides complement each other) and then, using the amino acid coding table, decode the order of amino acids that would result if this mRNA was translated. Note that this flow of information only goes one way. Protein cannot be turned back into RNA or DNA.